Covid-19 Analysis and Visualization using Plotly Express
Introduction: This project deals with creating dozens of bar charts, line graphs, bubble charts, scatter, and plots. The graph that will be made in this project will be of excellent quality. Envisioning COVID-19 will primarily be using Plotly Express for this project. The analysis and visualization enable people to understand complex scenarios and make predictions about the future from the current situation.
This analysis summarizes the modeling, simulation, and analytics work around the COVID-19 outbreak around the world from the perspective of data science and visual analytics. It examines the impact of best practices and preventive measures in various sectors and enables outbreaks to be managed with available health resources.
Tools and Technologies Used in the Project: Google Colab(Runtime type - GPU).
Requirements to Build the Project: Basic knowledge of Python, Basic understanding of graphs and charts, Data Visualization, Pandas, Numpy, Matplotlib and Plotly Express, Choropleth, Word Cloud.
Installation
Plotly Express is a high-level Python visualization library. It contains a function that generates complete graphs and plots. We can create complete graphs with just one line of code which will be creating lots of graphs across projects.
pip install plotly
Pandas is an excellent data analysis and manipulation tool for reading outliers.
pip install pandas
Matplotlib is an excellent data visualization library.
pip install matplotlib
Numpy is Numerical Python, we are using Numpy in this project for basic computing.
pip install numpy
Choropleth maps are used to plot maps with shaded or patterned areas which are proportional to a statistical variable.
pip install choropleth
Word Cloud is a visualization technique for texts
pip install wordcloud
A Step-by-Step Process to Implement the Project:
Task 1: Importing Necessary Libraries
#Importing Pandas and Matplotlib
#Data analysis and Manipulation
import pandas as pd
#Data Visualization
import matplotlib.pyplot as plt
# Importing Plotly
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.express as px
# Initializing Plotly
import plotly.io as pio
pio.renderers.default = 'colab' # To initialize plotly
Task 2: Importing the Datasets
Importing three datasets into this project
1. covid.csv- This dataset contains Country/Region, Continent, Population, TotalCases, NewCases, TotalDeaths, NewDeaths, TotalRecovered, NewRecovered, ActiveCases, Serious, Critical, Tot Cases/1M pop, Deaths/1M pop, TotalTests, Tests/1M pop, WHO Region, iso_alpha.
2. Covid_grouped.csv- This dataset contains Date(from 20-01-22 to 20-07-27), Country/Region, Confirmed, Deaths, Recovered, Active, New cases, New deaths, New recovered, WHO Region, iso_alpha.
3. covid+death.csv - This dataset contains real-world examples of a number of Covid-19 deaths and the reasons behind the deaths.
# Importing Dataset1
dataset1= pd.read_csv("covid.csv")
dataset1.head() # returns first 5 rows

# Returns tuple of shape (Rows, columns)
print(dataset1.shape)
# Returns size of dataframe
print(dataset1.size)
(209, 17) 3553
# Information about Dataset1
dataset1.info() # return concise summary of dataframe
<class 'pandas.core.frame.DataFrame'> RangeIndex: 209 entries, 0 to 208 Data columns (total 17 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Country/Region 209 non-null object 1 Continent 208 non-null object 2 Population 208 non-null float64 3 TotalCases 209 non-null int64 4 NewCases 4 non-null float64 5 TotalDeaths 188 non-null float64 6 NewDeaths 3 non-null float64 7 TotalRecovered 205 non-null float64 8 NewRecovered 3 non-null float64 9 ActiveCases 205 non-null float64 10 Serious,Critical 122 non-null float64 11 Tot Cases/1M pop 208 non-null float64 12 Deaths/1M pop 187 non-null float64 13 TotalTests 191 non-null float64 14 Tests/1M pop 191 non-null float64 15 WHO Region 184 non-null object 16 iso_alpha 209 non-null object dtypes: float64(12), int64(1), object(4) memory usage: 27.9+ KB
# Importing Dataset2
dataset2= pd.read_csv("covid_grouped.csv")
dataset2.head() # return first 5 rows of dataset2

# Returns tuple of shape (Rows, columns)
print(dataset2.shape)
# Returns size of dataframe
print(dataset2.size)(35156, 11) 386716
# Information about Dataset2
dataset2.info() ## return concise summary of dataframe<class 'pandas.core.frame.DataFrame'> RangeIndex: 35156 entries, 0 to 35155 Data columns (total 11 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Date 35156 non-null object 1 Country/Region 35156 non-null object 2 Confirmed 35156 non-null int64 3 Deaths 35156 non-null int64 4 Recovered 35156 non-null int64 5 Active 35156 non-null int64 6 New cases 35156 non-null int64 7 New deaths 35156 non-null int64 8 New recovered 35156 non-null int64 9 WHO Region 35156 non-null object 10 iso_alpha 35156 non-null object dtypes: int64(7), object(4) memory usage: 3.0+ MB
Task 3: Data Cleaning
Data cleaning is the process of altering, modifying a record set, correcting erroneous records from the database and identifying incomplete, incorrect or irrelevant parts of the data and then removing dirty data.
# Columns labels of a Dataset1
dataset1.columnsIndex(['Country/Region', 'Continent', 'Population', 'TotalCases', 'NewCases',
'TotalDeaths', 'NewDeaths', 'TotalRecovered', 'NewRecovered',
'ActiveCases', 'Serious,Critical', 'Tot Cases/1M pop', 'Deaths/1M pop',
'TotalTests', 'Tests/1M pop', 'WHO Region', 'iso_alpha'],
dtype='object')We don't need 'NewCases', 'NewDeaths', 'NewRecovered' columns as they contains NaN values. So drop these columns by drop() function of pandas.
# Drop NewCases, NewDeaths, NewRecovered rows from dataset1
dataset1.drop(['NewCases', 'NewDeaths', 'NewRecovered'], axis=1, inplace = True)
# Select random set of values from dataset1
dataset1.sample(5) 
Let's create a table through the table function already available in plotly express-
# Import create_table Figure Factory
from plotly.figure_factory import create_table
colorscale = [[0, '#4d004c'],[.5, '#f2e5ff'],[1, '#ffffff']]
table = create_table(dataset1.head(15), colorscale=colorscale)
py.iplot(table)
Task 4: Bar graphs- Comparisons between COVID infected countries in terms of total cases, total deaths, total recovered & total tests
Using one line of code, we will create amazing graphs using Plotly Express. Visualization can be done easily by moving the cursor in any plot, we can get label presence point directly by using the cursor. We can visualize and analyze the dataset with each aspect using the relation between the columns.
Primarily look at the country with respect to a total number of cases by top 15 countries only and color total cases and hover data as 'Country/Region', 'Continent'.
px.bar(dataset1.head(15), x = 'Country/Region', y = 'TotalCases',color = 'TotalCases', height = 500,hover_data = ['Country/Region', 'Continent'])
As the plot clearly shows the data for the top 15 countries, now again take the country with respect to the total number of cases from the top 15 countries, color the total deaths hover data as 'Country/Region', 'Continent' and analyze the visualization.
px.bar(dataset1.head(15), x = 'Country/Region', y = 'TotalCases',color = 'TotalDeaths', height = 500,hover_data = ['Country/Region', 'Continent'])
Let's analyze by coloring the total number of recovered cases
px.bar(dataset1.head(15), x = 'Country/Region', y = 'TotalCases',color = 'TotalDeaths', height = 500,hover_data = ['Country/Region', 'Continent'])
Visualize the same again by coloring the total number of tests.
px.bar(dataset1.head(15), x = 'Country/Region', y = 'TotalCases',color = 'TotalTests', height = 500,hover_data = ['Country/Region', 'Continent'])
The visualization could be as we have done with the top 15 countries with total cases, deaths, recoveries and tests. We can analyze the plot by looking at them.
Let's create a horizontal orientation plot with X-axis as 'TotalTests' and Y-axis as 'Country/Region' with passing parameter orientation="h" and color the plot by 'TotalTests'.
px.bar(dataset1.head(15), x = 'TotalTests', y = 'Country/Region',color = 'TotalTests',orientation ='h', height = 500,hover_data = ['Country/Region', 'Continent'])
Let's look at 'TotalTests' followed by 'Continent' and color the plot with 'Continent'.
px.bar(dataset1.head(15), x = 'TotalTests', y = 'Continent',color = 'TotalTests',orientation ='h', height = 500,hover_data = ['Country/Region', 'Continent'])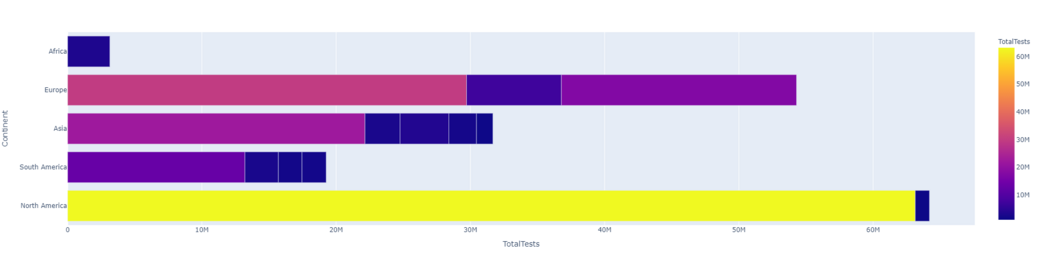
Task 5: Data Visualization through Bubble Charts-Continent Wise
Let's create a scatter plot and take a look at the continent's statistics, firstly look at the total number of cases by continent and take hover data as 'Country/Region', 'Continent'.
px.scatter(dataset1, x='Continent',y='TotalCases', hover_data=['Country/Region', 'Continent'],
color='TotalCases', size='TotalCases', size_max=80)
log_y= True, the histogram axis (not the returned parameter) is in log scale. The return parameter (n, bins), i.e. the values of bins and sides of bins are the same for log=True and log=False. This means both n==n2 and bins==bins2 are true
px.scatter(dataset1.head(57), x='Continent',y='TotalCases', hover_data=['Country/Region', 'Continent'],
color='TotalCases', size='TotalCases', size_max=80, log_y=True)
px.scatter(dataset1.head(54), x='Continent',y='TotalTests', hover_data=['Country/Region', 'Continent'],
color='TotalTests', size='TotalTests', size_max=80)
px.scatter(dataset1.head(50), x='Continent',y='TotalTests', hover_data=['Country/Region', 'Continent'],
color='TotalTests', size='TotalTests', size_max=80, log_y=True)
Task 6: Data Visualization through Bubble Charts-Country Wise
Let's take a look at the country-wise data visualization, first look at the continent with respect to the total number of deaths by top 50 countries only and color the total number of deaths and take the hover data as 'Country/Region', 'Continent'.
px.scatter(dataset1.head(50), x='Continent',y='TotalDeaths', hover_data=['Country/Region', 'Continent'],
color='TotalDeaths', size='TotalDeaths', size_max=80, log_y=True)
Now, the Country/Region with respect to the total number of cases for top 30 countries only and color the total number of cases and take the hover data as 'Country/Region', 'Continent'.
px.scatter(dataset1.head(30), x='Country/Region', y='TotalCases', hover_data=['Country/Region', 'Continent'],
color='Country/Region', size='TotalCases', size_max=80, log_y=True)
Now format the image of the country/region in relation to the total number of deaths. And do the same for the other aspects of COVID-19 from dataset1.
px.scatter(dataset1.head(10), x='Country/Region', y= 'TotalDeaths', hover_data=['Country/Region', 'Continent'],
color='Country/Region', size= 'TotalDeaths', size_max=80)
px.scatter(dataset1.head(30), x='Country/Region', y= 'Tests/1M pop', hover_data=['Country/Region', 'Continent'],
color='Country/Region', size= 'Tests/1M pop', size_max=80)
px.scatter(dataset1.head(30), x='Country/Region', y= 'Tests/1M pop', hover_data=['Country/Region', 'Continent'],
color='Tests/1M pop', size= 'Tests/1M pop', size_max=80)
px.scatter(dataset1.head(30), x='TotalCases', y= 'TotalDeaths', hover_data=['Country/Region', 'Continent'],
color='TotalDeaths', size= 'TotalDeaths', size_max=80)
It is clear from the result that they have a linear relationship between the total number of cases and the total number of deaths. That means more cases, more deaths.
px.scatter(dataset1.head(30), x='TotalCases', y= 'TotalDeaths', hover_data=['Country/Region', 'Continent'],
color='TotalDeaths', size= 'TotalDeaths', size_max=80, log_x=True, log_y=True)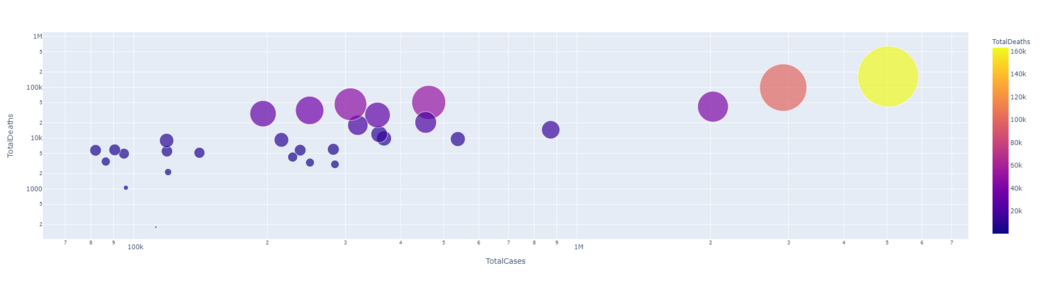
px.scatter(dataset1.head(30), x='TotalTests', y= 'TotalCases', hover_data=['Country/Region', 'Continent'],
color='TotalTests', size= 'TotalTests', size_max=80, log_x=True, log_y=True)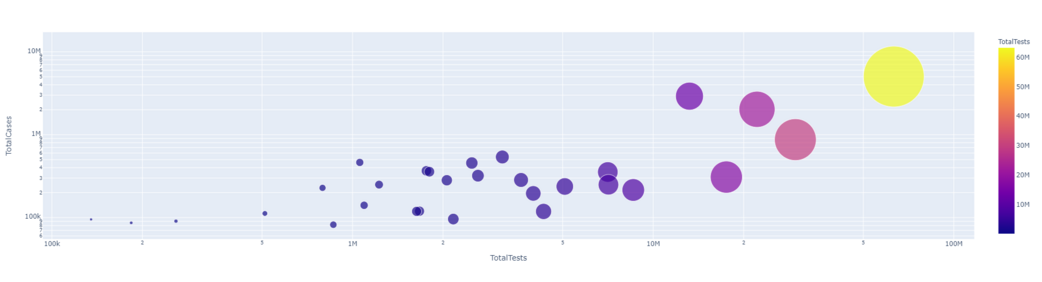
Task 8: Advanced Data Visualization- Bar graphs for All top infected Countries
In this task, we will explore covid-19 data using bar graphs and charts and use dataset2 as it has a date column.
px.bar(dataset2, x="Date", y="Confirmed", color="Confirmed", hover_data=["Confirmed", "Date", "Country/Region"], height=400)
The above graph we get as output which includes all countries with respect to recovered cases. we can imagine the exponential growth of corona cases by date. We can use log function for this to be more clear.
px.bar(dataset2, x="Date", y="Confirmed", color="Confirmed", hover_data=["Confirmed", "Date", "Country/Region"],log_y=True, height=400)
Let's imagine death instead of confirmation with the same and color it by date.
px.bar(dataset2, x="Date", y="Deaths", color="Deaths", hover_data=["Confirmed", "Date", "Country/Region"],log_y=False, height=400)
Task 9: Countries Specific COVID Data Visualization: (United States)
In this specific task we will analyze data from USA
df_US= dataset2.loc[df2["Country/Region"]=="US"]px.bar(df_US, x="Date", y="Confirmed", color="Confirmed", height=400)
Here we can clearly see how the confirmed cases increased in the United States with respect to time (January 2020 to July 2020).
Similarly, we can check the same for recovered cases, tests and deaths.
px.bar(df_US,x="Date", y="Recovered", color="Recovered", height=400)
Line Graph(United States)
Suchwise we can analyze the data in all the ways to generate the line graph for the same.
px.line(df_US,x="Date", y="Recovered", height=400)
px.line(df_US,x="Date", y="Deaths", height=400)
px.line(df_US,x="Date", y="Confirmed", height=400)
px.line(df_US,x="Date", y="New cases", height=400)
Bar Plot(United States)
px.bar(df_US,x="Date", y="New cases", height=400)
Scatter Plot(United States)
px.scatter(df_US, x="Confirmed", y="Deaths", height=400)
Task 10: Visualization of Data in terms of Maps
Using choropleth to visualize the data in terms of maps, with maps usually being the predominant way of visualizing the data. Since COVID-19 is a global phenomenon and so we look through and fix them in terms of wall maps. Ortho-graphics, rectangular and natural earth projection to visualize the data With dataset2 for the purpose as it has Dates column. It will look at the growth of Covid-19 (from Jan to July 2020) as in how the virus reached across the world.
Choropleth is an amazing representation of data on a map. Choropleth maps provide an easy way to visualize how a measurement varies across a geographic areal-Life
Project Application in Real choropleth map displays divided geographical areas or regions that are colored, shaded, or patterned in relation to a data variable.
Equi-rectangular Projection:
### Parameters used to map parameters= dataset, locations= ISOALPHA, color, hover_name, color_continuous_scale= [RdYlGn, Blues, Viridis...], animation_frame= Date
px.choropleth(dataset2,
locations="iso_alpha",
color="Confirmed",
hover_name="Country/Region",
color_continuous_scale="Blues",
animation_frame="Date")This will create an animation containing visualizations from January to July 2020. Playing this animation will make it more clear how the virus spread around the world. The darker the color, the higher the confirmed cases are.
px.choropleth(dataset2,
locations='iso_alpha',
color="Deaths",
hover_name="Country/Region",
color_continuous_scale="Viridis",
animation_frame="Date" )This code will create an animation of death cases by date. By playing this animation it will be shown how deaths increase around the world.
Natural Earth Projection:
Natural Earth projection is a compromise pseudo-cylindrical map projection for world maps.
px.choropleth(dataset2,
locations='iso_alpha',
color="Recovered",
hover_name="Country/Region",
color_continuous_scale="RdYlGn",
projection="natural earth",
animation_frame="Date" )By running the output, things start to become more clear about how the recovery rate changes with respect to the date.
Bar Graph Animation: Convert the bar graph into animation using the Dates column that is in dataset2.
px.bar(dataset2, x="WHO Region", y="Confirmed", color="WHO Region", animation_frame="Date", hover_name="Country/Region")When running the output, the animation will run from January to July 2020. It will show 6 different bar graphs, each continent has its own color representing the confirmed cases.
Task 11: Visualize text using Word Cloud
Visualize the causes of death due to covid-19, as covid-19 affects people in different ways, hence creating a word cloud to visualize the leading cause of covid-19 deaths. To visualize the text the steps need to be followed are-
- Used to convert data elements of an array into list.
- Convert the string to one single string.
- Convert the string into word cloud
Dataset3: This dataset contains real-world examples of a number of Covid-19 deaths and the reasons behind the deaths.
dataset3= pd.read_csv("covid+death.csv")
dataset3.head()
dataset3.tail()
dataset3.groupby(["Condition"]).count()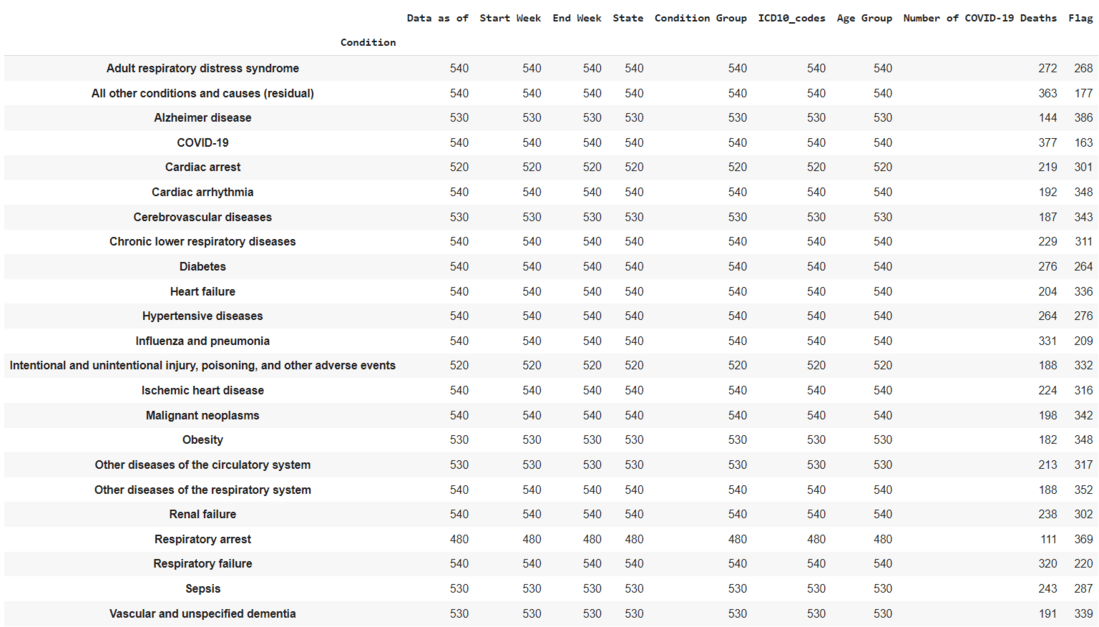
#import word cloud
from wordcloud import WordCloud
sentences= dataset3["Condition"].tolist()
sentences_as_a_string= ' '.join(sentences)
# Convert the string into WordCloud
plt.figure(figsize=(20,20))
plt.imshow(WordCloud().generate(sentences_as_a_string))
From the output, it can be clearly seen that the leading cause of death is Influenza Pneumonia.
column2_tolist= dataset3["Condition Group"].tolist()
# Convert the list to one single string
column_to_string= " ".join(column2_tolist)
# Convert the string into WordCloud
plt.figure(figsize=(20,20))
plt.imshow(WordCloud().generate(column_to_string))
We have converted the condition group to the list and stored the list in the variable "column_to_list".
column2_tolist= dataset3["Condition Group"].tolist()
# Convert the list to one single string
column_to_string= " ".join(column2_tolist)
# Convert the string into WordCloud
plt.figure(figsize=(20,20))
plt.imshow(WordCloud().generate(column_to_string))
Here we have converted the list into a single string and stored in a variable named "column2_to_string" by using .join().

Here, respiratory diseases are the major cause of death followed by circulatory diseases which are cardiovascular diseases.
Project Application in real life:
The analysis and visualization of COVID-19 help to make pandemic models more readily available.
Comments
Post a Comment